Chapter
2
The structure of four Ca2+ troponin C:
Insights into the Ca2+-switch
Note: This chapter first appeared in Structure [Houdusse, A., Love, M. L., Dominguez, R., Grabarek, Z. & Cohen C. : Structures of four Ca2+-bound troponin C at 2.0 Å resolution: further insights into the Ca2+-switch in the calmodulin superfamily. Structure 5, 1695-1711 (1997)][17]. It is presented largely as it was, and the text and references have been updated and corrected. My contributions to this work are as follows: I obtained all of the native and derivative crystals after a difficult search for the proper conditions, collected the first native and derivative data sets, and solved the derivative. With the assistance of Roberto Dominguez, the model was fitted to the resulting electron density map. I also collected the high resolution data at CHESS for the second crystal form, completed the refinement, produced 5 of the 6 figures for the paper, wrote the packing descriptions, and the second part of the Materials and Methods section as well. The contributions of the other authors are as follows: Roberto Dominguez finished the refinement of the first crystal form at high resolution, solved the second crystal form, and wrote the first part of the Materials and Methods section. Anne Houdusse produced figure 6 and wrote the description of the structural features, the conformational switch, the central helix comparison, and TnC/TnI interactions sections. Zenon Grabarek provided the protein, and many insights into the biochemistry of the system. Carolyn Cohen wrote the Biological Implications section, and provided guidance overall in the writing and editing.
2.1 Introduction
We have determined the structures of two forms of expressed rabbit Ca24+ -bound TnC to 2.0 Å resolution. The structures show that the conformation of the N-terminal lobe (N lobe) is similar to that predicted by the HMJ model. Our results also reveal, in detail, the residues involved in binding of Ca2+ in the regulatory N lobe of the molecule. We show that the central helix, which links the N and C lobes of TnC, is better stabilized in the Ca 22+-bound than in the Ca 24+-bound state of the molecule. Comparison of the crystal structures of the off and on states of TnC reveals the specific linkages in the molecule that change in the transition from off to on state upon Ca2+-binding. Small sequence differences are also shown to account for large functional differences between CAM and TnC. The two lobes of TnC are designed to respond to Ca2+-binding quite differently, although the structures with bound Ca2+ are very similar. A small number of differences in the sequences of these two lobes accounts for the fact that the C lobe is stabilized only in the open (Ca2+-bound) state, whereas the N lobe can switch between two stable states. This difference accounts for the Ca2+-dependent and Ca2+-independent interactions of the N and C lobe. The C lobe of TnC is always linked to TnI, whereas the N lobe can maintain its regulatory role - binding strongly to TnI at critical levels of Ca2+ - and in contrast, forming a stable closed conformation in the absence of Ca2+.
2.2.1 Overall structure description
The
overall conformation of the rabbit Ca 24+-bound
TnC molecule (Figure 2.1) described here from the study of two crystal
forms is generally similar to the well known crystal structure of Ca24+-bound
CAM [49]. The molecule consists of two lobes, each comprising two EF-hand
domains joined by a linker. Each EF-hand domain or helix-loop-helix motif
is analogous to the EF hand first described for parvalbumin [50]. In both
crystal forms of rabbit TnC, a Ca2+ ion is bound in the loop
of each of the four EF hands. The structure therefore consists of two open
lobes in which the angles between the incoming and outgoing helices of
each domain are between 90° and 100° (Table 2.1). In this open
state, the wide interior hydrophobic surface in each lobe is exposed. As
expected, the Ca24+-bound TnC structures
differ from the Ca 22+-bound
TnC structure [15,16] in the conformation of the N lobe. In the Ca22+-bound
TnC structure, the angles between the incoming and outgoing helices of
each domain are about 140° and, because the helices pack close to one
another, the hydrophobic surface is largely internalized leading to a closed
conformation. As found for the crystal structures of the Ca24+-bound
CAM [49] and that of the Ca 22+-bound
TnC, the inter-lobe linker adopts a helical conformation in both crystal
forms of rabbit TnC - the last helix of the N lobe (D helix) and the first
helix of the C lobe (E helix) together with the linker form a long helix
comprising nine turns. TnC has a three-residue insertion in the middle
of this inter-lobe linker that is absent in CAM; the central helix consists
of only eight turns in CAM. An additional distinct feature of TnC is the
twelve N-terminal residues (T1-Y10), forming the so-called "N helix" of
the N lobe, which are not present in CAM. The structures of the N lobes
of the two crystal forms of rabbit TnC are very similar. The molecules
differ, however, in the bending of the central helix as well as the detailed
conformation of the C lobe as discussed below. Moreover, we have found
that the structures of the N lobe of rabbit TnC are more similar to the
HMJ model [22] or to the crystal structure of the N lobe of the Ca24+-bound
CAM than to the NMR solution structures of the Ca2+-bound N
lobe of TnC [59] and the whole TnC molecule [60].
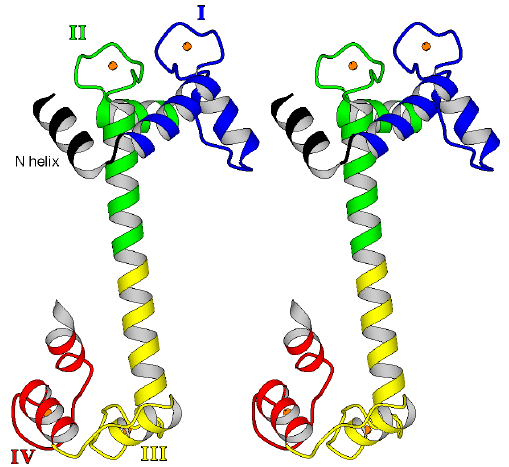
Figure 2.1
Stereo ribbon diagram of the overall fold of the four Ca2+-bound TnC. TnC has two lobes, each consisting of a pair of helix-loop-helix (EF hand) domains connected by a linker. The nomenclature is as follows: N helix is the N-terminal helix specific for TnC; domain I begins with helix A, followed by a loop, then helix B followed by a linker; domain II begins with helix C, followed by a loop, then helix D which continues into a helical linker, followed by helix E of domain III. Domains III and IV constitute the C lobe. A Ca2+ ion (sphere) is bound in each EF-hand loop of the structure, so that both lobes of TnC adopt an open conformation. The so-called central helix is composed of helix D, the inter-lobe helical linker and helix E. (Stereo diagram generated using the program MOLSCRIPT [87].)
| Table 2.1. | Interhelical angles | |||||||||||||||||||||||||||||||
| N-lobe TnC | A/B | A/D | B/C | B/D | C/D | ref. | ||||||||||||||||||||||||||
| open | 4Ca-TnC, X-ray (orthorhombic) | 97.0 | 116.4 | 119.2 | 34.3 | 98.9 | current | |||||||||||||||||||||||||
| open | 4Ca-TnC, X-ray (monoclinic) | 99.9 | 118.2 | 119.8 | 38.3 | 99.7 | current | |||||||||||||||||||||||||
| open | 2Ca-TnC N-lobe, NMR | 85.4 | 116.3 | 112.6 | 48.4 | 73.6 | [59] | |||||||||||||||||||||||||
| open | 4Ca-TnC, NMR | 78.2 | 95.6 | 125.6 | 60.7 | 94.3 | [60] | |||||||||||||||||||||||||
| closed | 2Ca-TnC, X-ray | 139.5 | 122.1 | 131.9 | 41.9 | 147.7 | [16] | |||||||||||||||||||||||||
| N-lobe CAM | A/B | A/D | B/C | B/D | C/D | |||||||||||||||||||||||||||
| open | 4Ca-CAM w/MLCK peptide, X-ray | 93.9 | 129.2 | 121.2 | 49.7 | 80.3 | [48] | |||||||||||||||||||||||||
| open | 4Ca-CAM, X-ray | 98.8 | 129.3 | 131.0 | 64.6 | 90.2 | [19] | |||||||||||||||||||||||||
| closed | apo-CAM, NMR | 140.2 | 135.2 | 132.5 | 42.9 | 130.6 | [57] | |||||||||||||||||||||||||
| closed | apo-CAM, NMR | 140.5 | 127.8 | 134.6 | 49.9 | 136.0 | [58] | |||||||||||||||||||||||||
| N-lobe RLC | A/B | A/D | B/C | B/D | C/D | |||||||||||||||||||||||||||
| open | myosin regulatory light chain | 98.8 | 129.3 | 131.0 | 64.6 | 90.2 | [67] | |||||||||||||||||||||||||
| C-lobe TnC | E/F | E/H | F/G | F/H | G/H | |||||||||||||||||||||||||||
| open | 4Ca-TnC, X-ray (orthorhombic) | 106.6 | 126.6 | 133.7 | 51.6 | 109.0 | current | |||||||||||||||||||||||||
| open | 4Ca-TnC, X-ray (monoclinic) | 120.9 | 121.7 | 121.8 | 36.6 | 117.0 | current | |||||||||||||||||||||||||
| open | 2Ca-TnC, X-ray | 106.4 | 118.8 | 132.5 | 34.7 | 112.9 | [16] | |||||||||||||||||||||||||
| open | 4Ca-TnC, NMR | 88.9 | 111.0 | 139.0 | 35.9 | 110.2 | [60] | |||||||||||||||||||||||||
| C-lobe CAM | E/F | E/H | F/G | F/H | G/H | |||||||||||||||||||||||||||
| open | 4Ca-CAM w/MLCK peptide, X-ray | 104.0 | 129.7 | 123.5 | 41.0 | 94.9 | [48] | |||||||||||||||||||||||||
| open | 4Ca-CAM, X-ray | 108.7 | 132.2 | 117.4 | 37.8 | 90.5 | [19] | |||||||||||||||||||||||||
| almost closed | apo-CAM, NMR | 137.5 | 156.9 | 145.4 | 32.7 | 135.0 | [57] | |||||||||||||||||||||||||
| almost closed | apo-CAM, NMR | 155.0 | 162.1 | 142.8 | 34.0 | 131.3 | [58] | |||||||||||||||||||||||||
The
angles between the helices were calculated after fitting a vector to the
backbone atoms of each helix using a least squares procedure. In the case
of a curved helix, ten residues closest to a calcium loop were chosen for
the least squares fitting.
2.2.2 Additional features of the structures
The N lobe of TnC
The
open conformation in the N lobe of the molecules is very similar in both
crystal forms of rabbit TnC as shown by the low root mean square (rms)
deviation (0.744 Å) of the Ca atoms for
the whole N lobe (between residues T1 and K84; Figure 2.2). (Note that
all residue numbers in the text refer to rabbit TnC
which differs by three residues from the avian TnC sequences.) This open
state differs significantly from that of the N
lobe
of Ca2+-bound TnC as described in recent NMR studies [59,60].
The rms deviations of the Ca
atoms for 44 residues of the four helices of the N
lobe (namely E13-M25 for helix A; V36-L46 for helix B; K52-E61 for helix
C; and F72-R81 for helix D) are 1.8,1.6 and 0.5 Å, when one compares
the C2 crystal form of rabbit TnC with the solution structure
of the N lobe of TnC, the solution structure of the Ca2+-bound
whole TnC and the orthorhombic crystal form of rabbit TnC, respectively.
These rms deviations derive from the fact that the lobes modeled from the
NMR spectroscopic studies are more open. A key difference in the results
from the NMR spectroscopic and crystallographic studies is found in the
position of the C helix. The interhelical angles A-B, B-D and C-D are also
different (Table 2.1). The conformation of the loops as well as that of
the BC linker, which are not well defined in the NMR solutions structures,
are even more divergent. The basis for this difference is not yet clear,
but it may result from the limited resolution of the NMR spectroscopic
data. The fact that the N lobe dimerizes when isolated does not seem to
be the explanation, because a study of the whole TnC (carried out in the
presence of trifluoroethanol to prevent the dimerization) gave a similar
structure to that of the N-lobe fragment [59,60]. The conformation of the
N lobe of TnC revealed by our results is very like that in both the HMJ
model and Ca24+-CAM. The rms differences
for the same 44 atoms of the N lobe of rabbit TnC in the C2 space group
are 1.1, 1.25 and 1.15 Å when compared to the HMJ model, the crystal
structure of the N lobe of Ca24+-bound
CAM and the crystal structure of Ca 24+-bound
CAM in complex with a peptide [48], respectively.
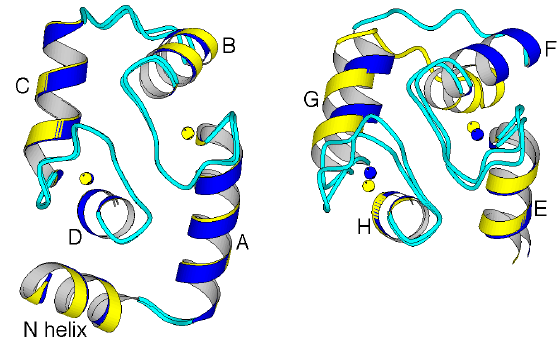
Figure 2.2
Comparison of the N and C lobes of rabbit
Ca 24+ -bound TnC in the monoclinic
(light) and orthorhombic (dark) crystal forms. (a) The N lobes are very
similar in both crystal forms. (b) In contrast, the conformations of the
C lobes differ, because the F-G pair of helices is oriented slightly differently
relative to the E-H pair of helices. The C lobe is therefore more open
in the orthorhombic form. This difference, produced by the intermolecular
interactions, shows that the C lobe of TnC is more flexible than the N
lobe.
The C lobe of TnC
Comparison
of the structures of the C lobe of rabbit TnC in the two crystal forms
shows that they differ significantly in the degree of opening of the lobe
(Figure 2.2). The rms difference between the Ca
atoms of all residues from E93-M154 of the two crystal forms is 1.7 Å.
In the C2 crystal form, the lobe is much less open than in the P212121
crystal form. The open C lobe of avian Ca22+-bound
TnC [15,16] is in a position that is intermediate between the
C lobes of the two crystal forms of rabbit TnC (see inter-helical angles
E-F, F-G, F-H and G-H in Table 2.1). Superimposition of the C lobes of
avian TnC onto the C lobes of the two crystal forms of rabbit TnC gives
rms differences in the Ca
atoms
of 1.4 Å for the C2 crystal form and 1.2 Å
for the P212121 crystal form. The main
difference between these three conformations is in the position of the
pair of helices F and G relative to that of the E and H helices; the orientation
of helix F diverges the most (see Table 2.1). The
conformations of the C lobes found in the two crystal forms of rabbit TnC
diverge the most from that of the C lobe in Ca 24+-bound
CAM [49]. Superimposition of the C lobe of Ca24+-bound
CAM onto that of avian TnC and that of rabbit TnC
in the P212121 and the C2 crystal forms
gives rms differences in the Ca
atoms
(between residues E93-M154) of 1.1, 1.3 and 1.6 Å, respectively.
Analysis of the interactions among the four helices of this open C lobe
shows no significant difference in the three TnC structures.
This finding is related to the fact that the turns
of the helices that interact with one another are close to the loop region
and make similar contacts regardless of their orientation. In contrast,
the turns of the helices which differ most correspond to those at the tips
of the lobe, which do not interact with one another. These tip regions
are involved in very different interactions with symmetry-related molecules
in these three crystal forms of TnC, which accounts for differences in
the position of the F and G helices and therefore in the amplitude of the
opening of the lobe. The binding of Ca2+ ions in the two EF
hands of the C lobe, therefore, does not require a very precise positioning
of the helices of the lobe. These conformational differences in the C lobes
of TnC in different packing environments suggest that this lobe possesses
a rather large internal flexibility. In the TnC-TnI complex, the C lobe
could easily adopt the open conformation that allows a best fit between
the two molecules.
Central helix
An unusual feature of the crystal structures of TnC is the nine-turn central helix (including the D and E helices), in which three solvent-exposed turns do not make interactions with either lobe of the molecule. (CAM has a similar helix with eight turns). We would expect that this part of the helix is only marginally stable in solution, and indeed NMR spectroscopic studies show that about two turns (residues M83-S91) in this region tend to unwind [60]; CAM is also unstable in the analogous region (residues M76-S81) [57,58]. Even in the crystal structures, there is evidence for some flexibility of this helix, although the B factors are not especially high in this region. In the two Ca 22+-TnC structures [15,16] and in the P212121 form of the rabbit Ca 24+-TnC, the helix is similarly bent. But in the C2 crystal form of rabbit Ca24+-TnC, due to specific packing contacts, it is bent differently, and this bending alters the relative positions of the two lobes of TnC (Figure 2.3). It is easy to picture that specific interactions of each lobe of TnC with different parts of TnI in the complex would be promoted by the flexibility of the central helix.
There is a major difference, however, in the stabilization of the central helix when one compares the closed [15,16] and open forms of TnC (either of structures of rabbit TnC described in this study). In the closed state of the N lobe, the B and C helices, as well as the linker between them, make numerous van der Waals contacts (about 40) as well as eight specific hydrogen bonds with the central helix, in the D-helix region between residues M78-D86. These linkages, which do not occur in the open state of the lobe, are the only ones that stabilize the helical turn between residues K84-D86. Thus, the central helix in the Ca2+-bound state of TnC has an additional solvent-exposed turn, which would enhance pliancy between the lobes in the on state. Note that in CAM, this helix is also more stable in the absence of Ca2+ [57]; it has been shown in solution, in the absence of Ca2+, that this region is helical for about one third of the time. The difference in stabilization of the central helix in the Ca2+-free and Ca2+-bound conformations accounts for the early observation that the K84-E85 and K88-G89 peptide bonds are readily hydrolyzed by trypsin in the presence of Ca2+, but not in its absence [61].
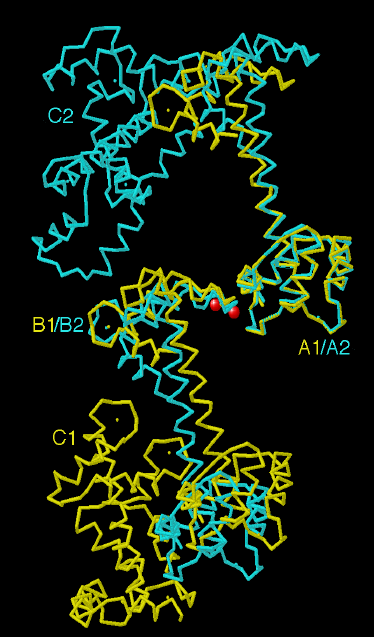
Figure 2.3
There is only one set of common interactions
around the TnC molecule in both the monoclinic and orthorhombic forms.
This involves two Ca2+ ions (spheres) found at the interface
between the C helix of the N lobe of one molecule (molecules A1 and A2)
and the FG linker of the C lobe of a neighboring molecule (molecules B1
and B2). The N lobes of molecule A1 (monoclinic) and A2 (orthorhombic)
are overlayed to show that their conformation does not deviate significantly
in the two crystal forms. In contrast, the central helix and the orientation
of the C lobe differ significantly due to the different packing constraints
in the two crystal forms. An additional molecule for each crystal form
(molecules C1 and C2) is also represented.
Calcium binding sites
The C lobe of TnC has two conventional EF-hand Ca2+-binding loops, to which either Mg2+ or Ca2+ ions can bind. In the crystal structures of rabbit TnC, as well as that of avian TnC, these sites are occupied by a Ca2+ ion. The coordination geometry around the Ca2+ ion is that of a pentagonal bipyramid and the ligands comprise three carboxylates - D103(X), N105(Y) and D107(Z) in loop III; D139(X), N141(Y) and D143(Z) in loop IV - one peptide carbonyl group - Y109(-X); R145(-X) - one bidentate glutamate - E114(-Z); E150(-Z) - and one bound water molecule (-Y). The first aspartate of loops III and IV (D103 and D139) and the water molecule occupy the axial positions of the pentagonal bipyramid. The conformation of the loop around the Ca2+ ion is very similar in sites III and IV of TnC and is also found in all four EF-hand domains of Ca 24+-CAM ([49]; Figure 2.4).
The N lobes in the refined structures of rabbit Ca 24+-TnC in the two crystal forms are very similar and allow us to describe in detail the coordination around the Ca2+ ions bound specifically in EF-hand domains I and II. The third ligand (Z) of these EF-hand loops differs from that in conventional EF-hands - rather than an aspartate there is a glycine (G31) in EF-hand loop I and a serine (S67) in EF-hand loop II. As in conventional EF-hand loops, however, the coordination around the Ca2+ ions is pentagonal bipyramidal and the geometry of the loops is similar to that found in conventional EF-hands (Figure 2.4). In EF-hand loop I, a water molecule substitutes for the missing aspartate, whereas in EF-hand loop II the serine binds directly to the Ca2+ ion. The other ligands, as in a conventional EF-hand, include two monodentate aspartates - D27(X) and D29(Y) for EF-hand I; D63(X) and D65(Y) for EF-hand II - one peptide carbonyl group - D33(-X); T69(-X) - one bidentate glutamate (E38(-Z); E74(-Z) - and one bound water molecule (-Y). In order to compensate for the short sidechain of S67 compared to that of an aspartate, the mainchain of the G66-S67-G68 adopts a slightly different conformation allowing the Ca of S67 to be about 0.6 Å closer to the Ca2+ ion than that of the aspartate in this position in a conventional EF-hand. A hydrogen bond between the sidechains of S67 and T69 also stabilizes the position of the serine bound to the Ca2+ ion in loop II. In domain I, the unusual water molecule bound to the Ca2+ ion is also stabilized by a hydrogen bond with D33. This aspartate residue, for which the position is homologous to that of T69 in loop II, is conserved among all skeletal TnCs. An aromatic or hydrophobic sidechain is found most commonly at this position in conventional EF-hands. This specific residue of TnC might therefore play a role in increasing the affinity of this loop for a Ca2+ ion both by adding a negative charge to the loop and by stabilizing the position of the water molecule which substitutes for the missing aspartate.
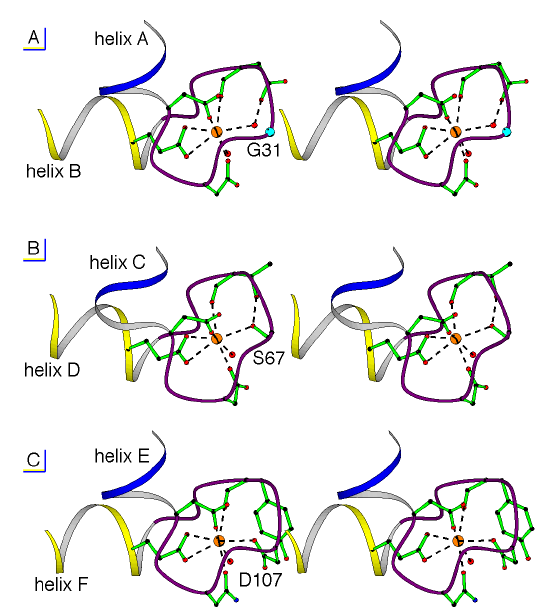
Figure 2.4
Structure of the Ca2+-binding site in EF-hand domain I (a), domain II (b) and domain III (c) showing the coordination around the Ca2+ ion (sphere). The Ca2+-ligands belong to a 12-residue loop located between an incoming helix and an outgoing helix, which are oriented roughly perpendicular to one another. The amino acid sequence in the loop of domain III (c) corresponds to a conventional binding site for an EF hand, whereas the third Ca2+-ligand of the loop in domain I (a) is a glycine (G31) and in domain II (b) is a serine (S67). The pentagonal bipyramid geometry of the ligands around the Ca2+ ion is, however, very similar in the three loops. A water molecule (sphere) plays the role of the missing aspartate in domain I, whereas a small change in the conformation of the loop near S67 allows the serine hydroxyl group to directly participate in the binding of the Ca2+ ion.
Unlike CAM, TnC possesses two functionally different lobes. The C lobe can bind both Mg2+ or Ca2+ ions with high affinity (Kd [Ca2+] ~ 10-7 M; Kd [Mg2+] ~ 10-3 M), whereas the N lobe is specific for Ca2+ ions in the presence of mM concentrations of Mg2+ because it binds these ions with lower affinity (Kd [Ca2+] ~ 10-5 M; Kd [Mg2+] ~ 10-1 M) [62]. This difference in cation affinity is directly related to the function of each lobe. Cations are always bound to the C lobe in vivo, so it is plausible that this lobe binds to TnI with conserved linkages in both the off and on states of the troponin complex and that it plays essentially a structural role in the complex [56]. By contrast, the N lobe which binds Ca2+ but not Mg2+ ions in vivo assumes one of two distinct conformations depending on the concentration of Ca2+ ions.
In
order to account for the specificity of the first and second EF-hand loops
for Ca2+, one should note that the geometry of these loops is
not changed by the fact that an aspartate is missing in the third position:
the loops remain as large as those in a conventional EF hand and would
therefore bind Ca2+ ions preferentially to Mg2+.
As noted above, the affinity for Ca2+ in these two EF hands,
however, is very weak, so that at physiological Mg2+ concentrations,
the binding of Mg2+ ions does not occur. The notion that the
N lobe is more selective for Ca2+-ions than the C lobe, is in
fact simply due to the difference in affinities of these lobes for Ca2+
ions and the ambient concentrations of Mg2+ ions. The structural
basis for cation affinity will be described in a later section.
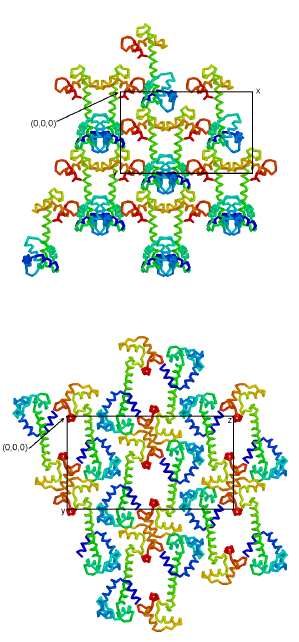
Crystal packing
The two different crystal forms of TnC allow an analysis of the effects of different packing interactions (Figure 2.5). In both forms, there are few packing interactions with the central helices and large solvent channels run roughly perpendicular to their axes. The hydrophobic patch in the interior of the lobes is fully exposed to these solvent channels. In the monoclinic crystal form, TnC is roughly oriented, N lobe to C lobe, along the twofold axis in accordance with the C2 packing constraints. This results in an axial polarity of molecular orientation that is not possible in orthorhombic crystals, in which the higher symmetry results in opposite orientations for symmetry related molecules. As a consequence, the central helix of each molecule interacts with different lobes in the two crystal forms. In the orthorhombic crystal, the central helix interacts with the B helix of a symmetry-related molecule, whereas in the monoclinic crystal the central helix interacts on the opposite side with EF-hand loop IV of a symmetry-related molecule (Figure 2.3). Despite the differences in packing interactions, the strongest set of interactions is common to both crystal forms. Two Ca2+ ions bridge D56 and E60 of the C helix of one molecule with the FG linker near E124 of a symmetry related molecule. Seven ligands, including three water molecules, are arranged around each of the two Ca2+ in a pentagonal bipyramid. In each crystal form, an additional Ca2+ ion is also involved in the interactions between symmetry-related molecules, although the location of these cations is different in the two forms.
Figure 2.5
TnC packs differently in the two crystal forms. In the monoclinic C2 crystal form (a), TnC is roughly oriented, N lobe to C lobe, along the twofold axis in accordance with the C2 packing constraints. This results in a polarity of direction, which is not possible in crystals with P212121 symmetry. In the orthorhombic P212121 crystal form (b), the molecules show opposed orientations due to higher symmetry constraints. As a result the packing interactions are very different for the two crystal forms and the molecules adopt somewhat different conformations. In both cases, the molecules are tilted into and out of the page in an alternating pattern.
In both crystal forms, the presence of two Ca2+ ions bound by the acidic sidechains of D56 and E60 of the C helix suggests a propensity of this side of the C helix to be associated with positively charged neighbors. Moreover, these residues are specific for the TnC molecule; the homologous residues in CAM are Q49 and N53. This feature of the surface of the N lobe might be related to the binding of TnI. Another feature concerns possible differences in flexibility of the different parts of the TnC molecule; the C lobe and central helix adopt different conformations in the two crystal forms, whereas the N lobe adopts a similar conformation in these forms (see above). In order to determine whether this finding is the result of a greater internal flexibility of the C than the N lobe, or if it is caused by the crystal packing, we have examined the crystal contacts of each of the lobes in the two crystal forms. The contacts made with symmetry-related molecules that are common to the two crystal forms are those between the N and C lobes through the bridging Ca2+ ions (as described above). This finding indicates that there is no preferential conservation of packing forces around the N lobe. In fact, the total number of contacts made by the N lobe with symmetry related molecules is significantly different in the two crystal forms (85 contacts in the orthorhombic space group and 62 in the monoclinic), whereas the number of contacts (~45) made with symmetry-related molecules by the C lobes are the same in the two crystal forms. It therefore appears that the open conformation of the C lobe is inherently more flexible than that of the N lobe. NMR spectroscopic experiments on CAM in the absence of calcium [57,58] have also revealed a similar difference in flexibility between the N and the C lobes of the molecule.
2.2.3 The conformational switch
The equilibrium between the open and closed conformations
Both lobes of CAM and the N lobe of TnC can adopt two major conformations - the open and closed states, depending on the presence or absence of Ca2+ ions in the EF-hand loops. A structure is not available for the Ca2+-free C lobe of TnC, but CD studies suggest that this lobe may be partially unfolded [63]. Specific sequence differences among these lobes are responsible for the differences in Ca2+-affinity, which appears to be related to the ability to switch between a closed and an open conformation in response to the Ca2+ ion concentration. The C lobe of TnC has a high binding constant for divalent cations [62], and it is always open in vivo. In contrast, the N lobe of TnC is easily closed and has the lowest binding constant for Ca2+ ions. The mechanism of the transition between these two states and the various factors which affect it will be described in detail, in order to relate the differences in the amino acid sequences of these lobes to differences in Ca2+ ion affinity.
The
conformation observed for the open state (see description above) is similar
in the N and C lobes of TnC and CAM because of their high sequence homology.
The closed state observed for the N lobe of TnC and CAM is also similar
[15,16,57,58]. The switching mechanism between the closed and open states
of the N lobes should therefore be similar in CAM and in TnC. The sequence
differences between these two lobes do not greatly affect how the switch
operates, but rather how easily the transition can be triggered by Ca2+
ions. By contrast, based on NMR spectroscopic studies, the conformation
of the C lobe of CAM in the absence of Ca2+ has been described
as "almost-closed" [64,54], because the packing of the helices in this
lobe differs from that in a true closed lobe. The inter-helical angles
E-H and F-H differ most significantly from those of a conventional closed
lobe (see Table 2.1). The mechanism of the switch in the C lobe of CAM
is therefore different from that in the N lobe. NMR spectroscopic studies
have also shown that this almost-closed conformation is not as stable as
the closed conformation of the N lobes of CAM or TnC, which are much less
flexible. The conformation of the C lobe of TnC in the absence of Ca2+
(as noted above) is also less stable than that of the N lobe. This difference
in stability of the Ca2+-free conformations in the C and N lobes
of both CAM and TnC is one of the basic factors that accounts for the difference
in Ca2+ ion affinity of these lobes.
The switch mechanism
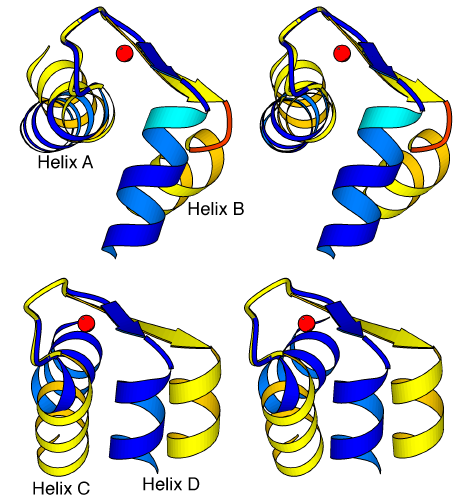
Comparison of the apo and Ca2+-bound forms of TnC allow us to identify the conserved and variable linkages that occur in the N lobe of this protein during the transition between the closed and open states. Moreover, the high-resolution crystal structures of TnC reveal these linkages in parts of the structures that are too flexible to be well-defined by NMR spectroscopic studies. The conformation of the mainchain for the first half of the Ca2+ binding loops (between residues D27-D33 in EF hand I and D63-T69 in EF-hand II) is conserved in the apo and Ca2+-bound states, so that Ca2+ binding affects only sidechain positions. In the apo-state at the center of both loops, a water molecule binds to the same four ligands that are involved in Ca2+-binding: the first two aspartates of the loop as well as the peptide carbonyl group and another bound water molecule. When a Ca2+ ion is substituted for this water molecule, a backbone conformational change occurs so that the 12th (-Z) residue of both loops (E38, E74) completes the coordination around the metal ion by binding via both oxygens of its carboxylate groups; the distance between the Ca of this residue and the location of the Ca2+ ion must then be reduced from ~10.5 Å to ~6.7 Å. As these glutamates are also the first residues of helices B and D, this movement directly affects the positions of the four helices of the lobe. As predicted by the HMJ model and found in the N lobe of TnC [59] and both lobes of CAM [57,58], the helices of the lobes rearrange in the transition so that the two helices of each domain become approximately perpendicular to one another. Although the helices move during this transition, the interactions between the two helical pairs A-D and B-C are conserved (see Table 2.1). Also, the N-terminal helix found only in TnC does not affect the transition, because it only interacts with the A-D pair of helices in a conserved manner in both states of the lobe. The interactions that change during the transition from a closed to an open form are therefore those between the A-B, B-D and C-D helices, which are for the most part hydrophobic interactions, as well as the more specific interactions occurring between the BC linker of the lobe and the D helix.
Comparison of the high resolution crystal structures of the apo and Ca2+-bound forms of TnC reveals the precise location of the hinges in the N lobe, which allow the helices to change their orientation in the two different states. A small hinge region allows helix A (or helix C) to reorientate slightly relative to the first seven residues of the following Ca2+-binding loop, and a larger hinge region allows helix B (or helix D) to markedly change its orientation relative to the Ca2+-binding loop (Figure 2.6). But the mechanism that operates the hinges is different in the two EF hands of the N lobe. The small hinge near helix A involves residues F26 and D27, which form a 310 turn of helix A in the apo-state but form a regular turn of an helix in the presence of the Ca2+ ion. This transition therefore requires the loss of two hydrogen bonds and the creation of three new ones for this last turn of helix A. In contrast, the last turn of helix C is conserved in both the apo and Ca2+-bound states. Here, the hinge encompasses the last two residues (V62 and D63) of the helix, and it causes the loss of one hydrogen bond of helix C during the transition from the apo to the Ca2+-bound state (i.e. the bond between the carbonyl group of I58 and the peptide NH group of V62). The differences in the conformational changes in the large hinges near helix B or helix D are even more pronounced. Near helix B, the hinge involves not only the residues of the strand - G32, I34 and S35 - but also residues V36, K37 and E38 which define the first turn of helix B only when a Ca2+ ion is bound. Three hydrogen bonds therefore form in the transition between the apo and Ca2+-bound states. In contrast, the hinge near helix D involves for the most part one residue (I70) of the preceding strand. The movement of helix B is
therefore
much greater than that of helix D due to the conformational change in the
first turn of helix B (residues V36-E38). The first and last hydrogen bonds
of the sheet are lost during the transition from the apo to the Ca2+-bound
state. The sheet is then less extended and less twisted when Ca2+
is present than in the apo state; and this allows the reorientation of
helices B and D. This knowledge of the detailed mechanism of the switch
provides a structural explanation for the cooperativity of Ca2+-binding
in the two EF-hands of a lobe. Not only do the helices move so that the
interactions between the helical pairs A-D and B-C are conserved (HMJ model),
but the concomitant shortening and twisting of the sheet in effect allows
one EF hand to sense the binding of a Ca2+ ion in the other
one.
Figure 2.6
Conformational switch in the EF hands of the N lobe of TnC. Stereo diagram of the EF-hand domains I (a) and II (b) in the Ca2+-free (light) and Ca2+-bound (dark) structures of the N lobe of TnC. The helices of a domain are reoriented during the transition by conformational changes occurring in two hinge regions. The mechanism of the hinges is different, in EF-hands I and II, however. In particular, the hinge is more extended near helix B, rather than near helix D; this is because in EF-hand I, it involves not only residues from the sheet but also residues T36-E38, which define an additional turn of helix B when Ca2+ is bound.
Comparison with CAM
How
does this switch mechanism in the N lobe of TnC relate to what is known
for CAM? A description of the hinges at the end of the Ca2+-binding
loops (near helices B and D in the N lobe and near helices F and H in the
C lobe) has been derived from an NMR spectroscopic study of apo-CAM [57].
The hinges at the beginning of the loop (next to helices A, C, E and G),
however, were not described, presumably because of the lack of resolution
for the Ca2+-binding loops (in particular for the C lobe. The
hinge near the B helix (which we have described as the formation of an
additional turn) had previously been pictured in CAM as undergoing a transition
from a "kinked" to a "regular" geometry during the binding of Ca2+
ions. The mechanism of the switch is in fact very similar for the N lobe
of CAM and TnC, but two small differences are found: firstly, the last
turn of helix A in CAM (where the small hinge of EF-hand I occurs) stays
in a 310 conformation in the presence of Ca2+, rather
than becoming a regular turn of an helix; secondly, the large hinge in
EF-hand II occurs at the beginning rather than at the end of the strand
(residues N60 and G61), which is moreover less extended in apo-CAM than
in apo-TnC. This difference could be related to the presence of a proline
(P66) in CAM rather than a glutamate (E73) in TnC at the beginning of helix
D. In contrast, the mechanism of the switch differs in major ways between
the C and the N lobes of CAM [57]. In the first EF hand of the C lobe,
the large hinge near helix F involves only residues I100 and S101 at the
end of the sheet. In the N lobe, the hinge is more extended because it
also involves the unwinding of the last turn of helix B upon the loss of
Ca2+ ions. This difference in the stability of helices F and
B is probably due to the following sequence differences: in CAM, helix
F has two alanine residues in a row (A102 and A103) and an arginine residue
at position 106, whereas the equivalent residues in helix B are V36-K37
and G40 in TnC and T29-K30 and G33 in CAM. Consequently, the movement of
helix F cannot be as large as that of helix B. These sequence differences
may be one factor affecting the conformational differences between the
N lobe (closed) and the C lobe (almost-closed) of CAM. Another factor may
be the small number of interactions between the FG linker and the H helix
in the C lobe which contrasts with the multiple interactions present in
the N lobe (see below). Differences also occur in the second EF hand of
these lobes. In the N lobe, the hinges of this EF hand do not involve any
major conformational change in the secondary structure of the helices.
In contrast, the G and H helices of the C lobe are somewhat destabilized
in the apo-state: one turn of helix G unwinds upon the removal of Ca2+
from the loop because of the loss of two hydrogen bonds (E123O-E127N and
I125O-D129N), and helix H is also shorter by one residue in the apo-state
because of the loss of one hydrogen bond (Y138O-V142N). Although this part
of the structure is not as well defined in the NMR solution structures
of apo-CAM, these same differences have been found in two studies [57,58].
Note that the occurrence of the almost-closed conformation in the C lobe
of CAM might be important, because CAM is predicted to bind to a variety
of target proteins with a semi-open conformation in the absence of Ca2+
[53,64,54]. In this case, fewer interactions would be disrupted in the
transition from an almost-closed to a semi-open conformation than would
occur in a transition from a closed state.
Sequence variations and affinity for Ca2+
The sequence differences described in the previous section might influence the affinity for Ca2+ ions in the different lobes of TnC and CAM, because they probably affect the switch mechanism. In addition, three other kinds of sequence variation in the lobes are even more likely to modulate the action of the switch between the open and closed states: the precise sequence of the Ca2+-binding loops, the hydrophobicity of the residues in the interior of the lobes and the residues involved in specific interactions characteristic of the closed state of the N lobe of TnC. The substitution of an aspartate residue by a glycine in the first EF hand of the N lobe of TnC appears to be an important factor in accounting for the low affinity of this lobe for Ca2+ ions. The hydrophobicity of the internal surface of the open lobes is a common characteristic of TnC and CAM which allows them to have generally similar functional properties. The more hydrophobic the interior of the lobe, the lower its affinity for divalent cations that promote the lobe opening and exposure of the interior to the solvent. Inspection of the hydrophobic core residues of both the N and C lobes in the crystal structures of TnC and CAM shows that these lobes are essentially equivalent in hydrophobicity (see figure which is available as supplementary material with the Internet version of this paper). We have found a few sequence differences, however, which might affect the switch. Firstly, two hydrophobic residues are present in the N lobe of TnC (M45 and V62) which are in equivalent positions to the less apolar residues in the C lobe of CAM (N111 and A128) and in the C lobe of TnC (A121 and S138). These hydrophobic residues would tend to decrease the affinity of the N lobe for Ca2+ ions compared with that of the C lobes. Point mutation studies have indeed confirmed that the mutation M45A in TnC increases the binding of Ca2+ ions [65]. Secondly, a hydrophilic threonine residue (T122) at the end of helix F in the C lobe of TnC instead of a hydrophobic leucine found at this position in the C lobe of CAM (L112) and in the B helix of both N lobes (L39, L46) interferes with the closed conformation of the C lobe of TnC (and therefore it enhances the Ca2+ affinity of this lobe).
One of the most important factors that appears to decrease the affinity for Ca2+ ions in the N lobe of TnC compared with the lobes of CAM results from the greater stability of the closed state of this lobe, due to specific interactions between the BC linker and the D helix. The sequence on one side of this helix is indeed specific to TnC (R81-Q82-E85-D86) and allows eight hydrogen bonds to be made with residues Q48, T49, E54 and E61 of the linker in the closed state of the lobe. A large number of van der Waals contacts also occur because of the close proximity between the BC linker and the D helix. The CAM sequence for this helix (R74-K75-D78-T79) does not allow these bonds to be made with the corresponding residues of the linker (Q41, N42, E47 and E54). In apo-CAM, this linker makes very few contacts with the D helix [57,58]. In order to bind Ca2+ ions in the N lobe of TnC, the transition to an open conformation requires that all these bonds be disrupted. A study of point mutations in TnC has previously shown the influence of the interactions between the BC linker and the D helix [66] on the switch. Mutants E54K or E85K of TnC display a decrease in Ca2+ affinity, because in the absence of Ca2+ ions, a salt bridge occurs in these mutants, substituting for the hydrogen bond found in the wild-type TnC. In the case of the C lobe, helix H is shorter than helix D so that specific interactions with the FG linker (equivalent to those described above for the D helix and the BC linker in the N lobe) cannot occur in the closed state. This finding may account for the higher Ca2+ affinity of the C lobe than in the N lobes of both TnC and CAM.
This analysis shows that the most important features of the N lobe of TnC that account for its particularly stable closed conformation are the specific sequences found in the Ca2+-binding loops as well as those found in the D helix that promote stabilizing interactions with the BC linker in the closed conformation of the lobe. These sequence differences promote the optimal design of each lobe for its functional role: the C lobe of TnC adopts an especially stable open conformation in the presence of divalent cations, whereas the N lobe has an especially stable closed conformation which switches to the open state only at a critical concentration of Ca2+ ions. Both lobes of CAM can switch states more readily than the N lobe of TnC.
2.2.4 Comparison of the central helix in TnC and CAM
In order to infer where the least stable region in the central helix of TnC might be located, we have compared this region to that in two other members of the CAM superfamily: CAM and the regulatory light chain (RLC) of scallop myosin, the structures of which have been determined in complexes with their targets [48,67,68]. Although CAM and the RLC have equal number of residues between the N and C lobes, those that define the interlobe linker in each complex are not analogous. The D helix in CAM terminates one turn (3 residues) earlier than that of RLC, whereas the E helix starts one turn (4 residues) earlier (Table 2.2). The sequence of TnC in this region differs from CAM and the RLC by a three-residue insertion (K88-G89-K90) containing the only glycine of the TnC helix - probably the most flexible point in the helix. This three-residue insertion would be located in the RLC sequence between residues D84 and S85, in the middle of the linker. In the CAM sequence, however, this insertion would be located between residues D80 and S81, that is in a region which corresponds to the E helix (this helix starts at residue D78; [48]). From this analysis, one would conclude that there is a Ca2+ dependent flexibility in the central helix of TnC between residues K84-S91. This is the region homologous to that of the so-called interlobe linker in the RLC-heavy chain (HC) complex, but not really homologous to the interlobe linker in the CAM-peptide complex.
Table
2.2. Sequence alignment in the central helix / inter-lobe linker region.
| CAM/peptide | TnC | RLC/HC | |||
| ... | ... | ... | |||
| E67 | E74 | M71 | |||
| F68 | F75 | F72 | |||
| L69 | L76 | L73 | |||
| T70 | V77 | S74 | |||
| Helix D | M71 | M78 | I75 | ||
| ---------- | M72 | M79 | F76 | ||
| A73+ | V80 | S77 | |||
| R74+ | R81 | D78 | |||
| K75+ | Helix D | Q82 | Helix D | K79 | |
| linker | M76+ | ---- on state | M83 | ------------ | L80 |
| K77+ | K84* | S81+ | |||
| D78+ | E85* | G82+ | |||
| ---------- | T79 | ---- off state | D86* | T83+ | |
| Helix E | D80 | A87+ | D84+ | ||
| - | K88+ | linker | - | ||
| - | linker ? | G89+ | - | ||
| - | K90+ | - | |||
| S81 | S91+ | S85+ | |||
| E82 | E92+ | E86+ | |||
| E83 | ------------ | E93 | ------------ | E87 | |
| E84 | Helix E | E94 | Helix E | T88 | |
| I85 | L95 | I89 | |||
| R86 | A96 | R90 | |||
| E87 | N97 | N91 | |||
| A88 | C98 | A92 | |||
| Ö | Ö | Ö |
The
residues belonging to the inter-lobe linker in the CAM/peptide and RLC/HC
complexes (in red) are not homologous. The hypothetical Ca2+
dependent unwinding of the central helix of TnC is predicted to include
residues K84-E92 (* and +) in the presence of Ca2+ but residues
A87-E92 (+) in the absence of Ca2+.
Comparison of solvent-exposed residues of the central helix also reveals that the location of the region likely to unwind upon binding to the target protein differs in CAM and TnC. Residues R74-K77, which constitute the linker region in the Ca2+-CAM bound to the MLCK peptide [48], form a turn of the central helix that is exposed to the solvent in the Ca2+-bound structure of CAM [49]. In contrast, in the Ca2+-bound structures of rabbit TnC (this study), the corresponding residues (R81-K84) interact with the helices of the N lobe (including the interactions via two specific hydrogen bonds formed by R81. The most important stabilizing interactions for this part of the central helix come from the N-terminal helix - the first ten residues of the TnC sequence, which are absent in the CAM sequence. As discussed previously, the central helix is even more protected in the Ca2+-free form of TnC, due to specific interactions with the BC linker which stabilizes an extra turn of helix between residues K84 and D86. The region of the TnC central helix, homologous to that of CAM, is therefore even less likely to unwind in the absence of Ca2+. In fact, a similar difference in the flexibility of the central helix is found in CAM, depending on the level of Ca2+ ions. Both NMR spectroscopic studies of apo-CAM [57,58] and modeling studies of apo-CAM-target-peptide complexes [54] show that the region that unwinds is M76-E82, rather than R74-K77 as in the Ca2+-CAM-peptide complex [48]. Taken together, these results show that, although the sequences of CAM and TnC are very similar in the region which unwinds in the CAM complex, the flexibility of the central helix is probably located in different regions for the two molecules and is also likely to be different in the Ca 22+- and the Ca24+-forms of TnC.
Mutation studies have been reported which support these inferences. In an attempt to induce some CAM-like functions in a TnC molecule, several mutants of TnC have been engineered [69]. Mutants in which either region specific to TnC (i.e. the N-terminal helix or the K88-G89-K90 residues of the central helix) were deleted separately were unable to elicit CAM-like regulation, whereas mutants with both regions deleted were CAM-like in function. We would infer that the deletion of the N-terminal helix probably destabilized the central helix near residues R81-K84, whereas the deletion of the K88-G89-K90 residues resulted in a linker of both the proper length and rigidity in a region which was designed to maintain the helix. In the same paper [69], the authors reported a TnC mutant with CAM-like regulatory properties by replacing the TnC-specific sequence E85-DAKG- K90 in the DE linker by the CAM-specific sequence D78-T79-D80. Restoration of the CAM sequence in the DE linker was sufficient to induce complex formation with target proteins specific for CAM, although the interactions with the N-terminal helix had to be disrupted in order to form a linker. This is a rather surprising result, because none of the residues of the sequence D78- T79-D80 are involved in contacts with the target peptides [51,48,52]. As the mutant in which K88-G89-K90 was deleted could not induce CAM-specific function, we conclude that the flexibility in the DE linker of the two sequences RQMK-DTD-SEEE and RQMK-EDA-SEEE is sufficiently different to determine when a complex with CAM-specific target proteins can be formed. These experiments indicate that the flexibility in this central helix may be critical in order to obtain a specific function. They also indicate how small differences in the sequence can affect this feature of the helix. The sequences in this critical region appear to have evolved to fit the different functional requirements of these two closely related proteins.
2.2.5 TnC-TnI interaction
The Ca2+-dependent change in the interaction between TnC and TnI switches the thin filaments in striated muscle between the off and on states and ultimately regulates the onset and relaxation of tension. The TnC-TnI interaction has been the subject of extensive studies (reviewed in [70,71]). Although some general features are beginning to emerge, the precise mechanism of the regulatory switch is not yet known. Here, we review briefly some of the main findings in this area and attempt to explain them, whenever possible, on the basis of the structural properties of TnC revealed by this and previous work.
A number of studies show that there are Ca2+-independent and Ca2+-dependent components in the TnC-TnI interaction. The TnC-TnI complex therefore is stable in solution regardless of Ca2+ concentration, and the apparent binding constant is approximately 1000-fold higher (Ka ~ 109 M-1) when Ca2+ is present [72,73]. Moreover, although both lobes of TnC bind TnI, the N lobe does so only in the presence of Ca2+ [74]; thus, the N lobe has been pictured as having a regulatory role and the C lobe a structural role [56]. Although these two lobes are homologous in sequence and have very similar structures in the presence of Ca2+, we have shown above how small differences in their sequence may account for differences in their structural stability as well as Ca2+ affinity. The unusual feature of the C lobe of TnC is its inability to adopt a stable closed conformation in the absence of Ca2+. In contrast, the N lobe of TnC has two different, but well defined stable structures depending on whether or not Ca2+ is bound. It is the stability of the N lobe in the Ca2+-free closed conformation that prevents its binding to TnI. Thus, it appears that the structural constraints in the Ca2+-free structure of the N lobe and the lack of such constraints in the C lobe are responsible for the Ca2+-dependent and Ca2+-independent interactions with TnI in these lobes.
By analogy with CAM, the open lobes of TnC would be expected to bind to amphipathic helical segments of TnI (Note: It now appears that this CAM analogy is partially incorrect; see section TnC/TnI interaction mechanism in Chapter 3). Little is known, however, about the structure of the TnC-binding sites on TnI. Two regions of TnI can bind to TnC: the N-terminal residues 1-47 and the so called "inhibitory peptide" comprising residues 96-116, which reduces the ATPase activity in reconstituted actomyosin [28,29]. Biochemical studies have indicated that the N-terminal fragment of TnI interacts with the C lobe of TnC [75], and the recently determined crystal structure of TnC in complex with this peptide fragment reveals that it binds as an helix in the hydrophobic pocket of the open C lobe of TnC [43]. Thus, it appears that the N-terminal segment of TnI represents at least part of the structural site responsible for anchoring TnI to TnC. The structure of a portion of the inhibitory peptide bound to TnC (comprising residues 104-115) has been characterized using the transferred nuclear Overhauser effect technique [45]. In this complex, the peptide forms a turn of amphipathic -helix with a central bend induced by the two consecutive prolines (P109 and P110). From the functional properties of the inhibitory peptide, one would infer that it interacts with the N lobe of TnC. In fact, a photosensitive crosslinker on C57 in helix C of this lobe was found to crosslink to the C-terminal part of the inhibitory peptide, after the two prolines [32].
From this information, a model of the interaction between TnC and the inhibitory peptide has been proposed [71], using an analogy with the RLC-HC complex [67,68], in which a bend at residue Q825 of the HC is followed by an amphipathic helix bound in the open N lobe of the RLC. Such a model has plausible features because the hydrophobic core of the N lobe of Ca 24+-TnC could interact with the hydrophobic side of an amphipathic helix after residue P110 (residues L111, V114, A118 and M121); the other side of the helix having polar and basic residues (R112, R113, R115, M116, S117, D119 and K123) could potentially provide additional stabilization of the complex through the contacts with some residues at the rim of the lobe. We should note that the analogy between the TnC-TnI and RLC-HC complexes is consistent with our findings (described above), that is the N lobe of TnC adopts a conventional open conformation (in contrast to the results of recent NMR spectroscopic studies [59,60]). This model may aid in the design of new experiments, but one should note that results from short peptides may lead to an oversimplified picture of the TnC-TnI interaction.
As described earlier in this paper, although TnC and CAM are very similar in structure, small differences in their amino acid sequence lead to differences in the stability of the Ca2+-free state which account for differences in their regulatory mechanisms. The TnC molecule is always bound to the thin filament, whereas CAM binds reversibly to its targets. It is the flexibility of the Ca2+-free form of the C lobe of TnC that promotes its binding to TnI, regardless of the level of Ca2+ ions. In contrast, the stable closed conformation of the N lobe of TnC prevents the binding of this lobe to TnI in absence of Ca2+, although the two molecules are in proximity. The Ca2+-dependent transition between the closed and open conformations of this lobe is necessary and sufficient for the regulatory function of TnC. In the case of CAM, the two lobes must operate together when Ca2+ is released, so that both can bind to the target proteins in a Ca2+-dependent manner. Moreover, small differences between the two lobes of CAM in the apo-state allow them to operate differently and to adopt different conformations. As shown in the model of the complex of apo-CAM bound to an IQ motif [54], this property might be important for efficient binding of CAM to some specific target proteins in the absence of Ca2+.
2.2.6 Biological implications
Transient changes in the concentration of Ca2+ ions within the cytosol regulate a great variety of biological processes. The effect of calcium is mediated by proteins that specifically bind calcium and assume a new conformation which, in turn, allows them to interact with and modulate the function of their respective target proteins. Troponin C (TnC) and calmodulin (CAM) are two such intracellular Ca2+ sensors. A common structural feature of this group of regulatory proteins is the presence of four helix-loop-helix Ca2+-binding domains called EF hands. These proteins are often dumbbell-shaped, consisting of two lobes (N lobe and C lobe), each of which contains two EF-hand domains; the lobes are connected by a flexible linker. In the absence of divalent cations, the lobes are closed and cannot bind target proteins. In the presence of Ca2+ ions, the lobes open exposing a hydrophobic interior which can grip amphipathic target helices and so modify the function of the target proteins.
TnC is the Ca2+-sensing component of the troponin complex which binds to the thin filaments in skeletal and cardiac muscle; it acts as a trigger in muscle by switching on contraction at critical levels of Ca2+ ions. The target protein of TnC is the inhibitory component in the troponin complex, troponin I (TnI). The C lobe of TnC binds to TnI regardless of the level of Ca2+ ions and has therefore been called the "structural" lobe, whereas the N lobe is a bistable switch which binds to TnI only at a critical level of Ca2+ ions and is called the "regulatory" lobe. CAM, unlike TnC which has a very specific function and a unique target, is the most ubiquitous Ca2+-sensing protein and the most versatile. CAM binds many targets and in certain cases, such as non-conventional myosins, can do so even in the absence of Ca2+ ions. In previous crystallographic studies, neither CAM nor TnC had been visualized in both the "off" and "on" states. By comparing the structures of these two proteins, each in a different state, a model was proposed which shows how the hydrophobic-binding pocket becomes available when calcium is bound (the so called HMJ model). The model is supported by a variety of experimental data, including NMR spectroscopic studies that reveal the structures of the two states of both proteins. The detailed conformational changes that take place upon switching between off and on states in TnC and CAM, however, have not yet been described.
In this paper, we report the structure determination of two crystal forms of TnC in the four calcium-bound (Ca 24+) or on state to 2.0 Å resolution. This is the first detailed structure determination of TnC in the active four calcium-bound state. The results are in accord with the HMJ model and in addition enable us to show precisely which linkages change in the transition between the off and on states. We have also identified the residues involved in the binding of Ca2+ ions in the regulatory N lobe of the molecule, and have shown that the central helix in TnC is better stabilized in the Ca 22+-bound than in the Ca 24+-bound state of the molecule. The latter observation can be explained in terms of the numerous specific and non-specific interactions that are made in the closed (Ca2+-free) state of the N lobe, between the BC linker and the central helix. These interactions do not occur in the Ca2+-bound state, so that there is an additional solvent-exposed turn which would increase the flexibility between the two lobes. TnC is always linked to a part of TnI by its C lobe, and it grips TnI in the N lobe only at a critical level of Ca2+ ions. Because of the close proximity of TnC to TnI within the troponin complex, a stable closed N lobe of TnC is required to prevent its interaction with TnI in the absence of Ca2+ ions. Our results reveal how the unique linkages stabilizing the closed N lobe of TnC in the off state are essential for the switching function (between the on and off states) of the molecule. Unlike TnC, both lobes of CAM switch conformations at similar calcium levels, so that CAM binds readily and reversibly to its targets. Our results show how small sequence differences between TnC and CAM account for this difference in the properties of these molecules. The results also emphasize that only close inspection of high resolution structures can reveal the distinguishing functional characteristic of diverse members of the CAM superfamily.
2.3.1 Crystallization and data collection
The expressed mutant C98L of rabbit TnC studied here has the same functional properties and crystallizes under the same conditions as the wild-type protein. Two crystal forms of TnC in the four Ca2+ state were obtained: orthorhombic (P212121) with unit cell dimensions a = 32.34 Å, b = 57.55 Å, c = 102.13 Å and monoclinic (C2) with unit cell dimensions a = 83.19 Å, b = 51.78 Å, c = 53.07 Å, = 121.68°. Crystals of both forms were grown by seeding under identical conditions at 4°C from a mixture of 2 µl of reservoir buffer (50 mM HEPES pH 7.2, 10 mM CaCl2 and MPD 48%). The partial specific volume [76] is similar for both crystal forms (~2.6 Å3/Da), corresponding to the presence of one TnC molecule of 18 kDa in the asymmetric unit. A mercury derivative was prepared by soaking the orthorhombic crystals obtained from wild-type protein (i.e. containing a cysteine residue at position 98) for 20 hours in 1 mM mersalyl acid. One native and one mersalyl acid heavy-atom derivative data set were collected at 4°C to about 3 Å resolution using a CCD area detector mounted on an Elliot GX13 X-ray generator equipped with focusing mirrors (Table 2.3). A second native data set was collected to a resolution of 1.93 Å from the orthorhombic crystals at -160°C, using a MAR imaging plate detector mounted on an Elliot GX13 X-ray generator equipped with focusing mirrors. Finally, one native data set from the monoclinic crystal form was collected to a resolution of 2.0 Å at -160°C using synchrotron radiation at CHESS (beamline A1, l =0.914 Å). Data reduction was carried out with the programs DENZO [77] and SCALEPACK [78]. Table 2.3 summarizes the statistics of the diffraction. Structure determination, refinement and quality of the structures The crystal structure of the orthorhombic crystal form was determined at 3.0 Å resolution using a combination of single isomorphous replacement phases and density modification with the CCP4 program DM [79]. The position of a single mersalyl acid heavy-atom site was determined from a Patterson difference map between the native and derivative data sets. The heavy-atom parameters were further refined using the program MLPHARE [80]. The phasing power of the unique mersalyl acid heavy-atom derivative was 1.64. An electron-density map, calculated with 3.0 Å SIR phases (figure of merit 0.39) and further refined by solvent flattening and histogram matching with DM (figure of merit 0.65), revealed the molecular envelope. This envelope was used to fit a homology-based model of TnC in the four Ca2+-bound state, in which the open conformation of the N lobe was modeled based on the conformation of the C lobe. Fitting the model into the envelope was facilitated by locating the position of the heavy-atom ligand (C98). Solving the structure by molecular replacement probably failed because of the internal symmetry characteristic of this class of proteins, as well as the variability of conformations displayed by some parts of the molecule (for example the F helix of the C lobe). The initial crystallographic R factor of the homology model after manual fitting in the map was 60%. After 200 cycles of positional refinement with the program X-PLOR [81] using data to 3 Å resolution, the R factor fell to 38%. The refinement was then continued with the program ARP [82] using data to the highest resolution of 1.95 Å. A combination of restrained and unrestrained refinement with ARP produced new (3Fo-2Fc) electron-density maps that allowed major manual rebuilding of the model using the program O [83] running on a Silicon Graphics work station. The final refined model has an R factor of 18.9% and a Rfree of 25.3% for all data in the resolution range of 10-1.95 Å (Table 2.4). The model displays good stereochemistry, as judged with programs X-PLOR [81] and PROCHECK [84]. The rms deviations from ideal bond lengths and angles are 0.015 Å and 1.65°, respectively. Only a few exposed amino acid sidechains (Y10, E18, K84, E85, E113, D137 and E156) and the N-terminal (T1) and C-terminal (G157) residues show poor density definition. All mainchain dihedral angles of the non-glycine residues lie in allowed regions of the Ramachandran plot, with 95.8% in the most favored regions. The solvent structure was automatically modeled with the program ARP. New water molecules were added at positions where the difference electron density map was 4 and only in cases in which potential interactions with protein atoms were detected. At the end of the refinement, all water molecules with temperature factors higher than 40 Å2 were removed from the model. The average value of the temperature factors for protein atoms and water molecules are 15.6 Å2 and 25.8 Å2, respectively. The final model contains 181 water molecules.
The monoclinic form was solved by molecular replacement with program AMoRe [85], using the refined orthorhombic structure as a search model. Solutions were found for the N and C lobes, independently, using data from 8 to 4 Å resolution. The final correlation coefficient after fitting together the N and C lobe solutions was 46.6 (R factor of 48%) for all the data between 10 and 2.4 Å resolution. After one round of simulated annealing with the resolution extended to 2 Å, the crystallographic R factor fell to 29%. The refinement was continued and the solvent structure was modeled with the program ARP. The final refined model has a crystallographic R factor of 22% and an Rfree of 27%. The statistics of the refinement and model quality are listed in Table 2.4.
|
Table
2.3. Data collection statistics.
|
||||
|
|
|
|
|
|
|
Crystal
form
|
|
|
|
|
|
Temperature
of data collection
|
|
|
|
|
|
Number
of crystals collected
|
|
|
|
|
|
Resolution
range (Å)
|
|
|
|
|
|
Observed
reflections
|
|
|
|
|
|
Unique
reflections
|
|
|
|
|
|
Multiplicity
|
|
|
|
|
|
Completeness
(%)
|
|
|
|
|
|
Rmerge
(%)
|
|
|
|
|
|
Table
2.4. Refinement statistics.
|
||||
|
|
|
|||
|
Resolution
range (Å)
|
|
|
||
|
Number
of reflections
|
|
|
||
|
Sigma
cutoff
|
|
|
||
|
Completeness
(%)
|
|
|
||
|
Rfactor
(%)
|
|
|
||
|
Free
Rfactor(%)
|
|
|
||
|
Number
of protein atoms
|
|
|
||
|
Number
of Ca2+ ions
|
intermolecular contacts |
intermolecular contacts |
||
|
Number
of water molecules
|
|
|
||
|
Average
B-factor for protein atoms (Å2)
|
|
|
||
|
Average
B-factor for Ca2+ ions in EF-hand loops
(Å2)
|
|
|
||
|
Average
B-factor for Ca2+ ions in intermolecular
contacts (Å2)
|
|
|
||
|
Average
B-factor for water molecules (Å2)
|
|
|
||
|
rms
bond lengths (Å)
|
|
|
||
|
rms
bond angles (degrees)
|
|
|
||
|
rms
dihedral (degrees)
|
|
|
||
|
rms
impropers (degrees)
|
|
|
||
|
Global
G-factor
|
|
|
||
Accession
numbers The atomic coordinates of the refined TnC models have been deposited
with the Brookhaven Protein Data Bank [86], with the codes 1TN4 and 2TN4.
The coordinates can also be obtained by e-mail from CC at ccohen@binah.cc.brandeis.edu.
Supplementary
material
Supplementary
material available with the Internet version of this paper contains a stereo
diagram showing the N lobe of TnC (in wireframe representation) for both
the open and closed forms.
Note added in proof After this manuscript was accepted, a high resolution structure of the calcium-saturated N-terminal lobe of chicken troponin C was published (Strynadka, N.C., Cherney, M., Sielecki, A.R., Li, M.X., Smillie, L.B. & James, M.N. (1997). Structural details of a calcium-induced molecular switch: X-ray crystallographic analysis of the calcium-saturated N-terminal domain of troponin C at 1.75 Å resolution. J. Mol. Biol. 273, 238-255). This important paper complements our own study by presenting a detailed comparison of the calcium-binding loops as well as a model for the sequential binding of calcium to the two EF-hands of the regulatory lobe. The structures of the lobes are in good agreement with the current N lobe structures with an r.m.s.d of 0.420 Å. (There is some variability in the disposition of helix N, and it was excluded from this calculation.)